Kh. T. Kholmurodov, M. V. Avdeev, T. V. Tropin, A.V. Tomchuk, P. Grubovchak, E. V. Ermakova, A. I. Ivankov, T. Kondela, A. I. Kuklin, S. A. Kurakin, T. N. Murugova, V. V. Skoy, D. V. Soloviov, N. Kucherka
Our research includes molecular dynamics (MD) simulation of the structure and properties of new functional materials and nanosystems. Within the framework of the Topical Plan for JINR Research and International cooperation, we successfully carry out international collaborations: JINR-Japan: Nagoya, Keio-Yokohama, Waseda-Tokyo Universities; JINR-India: National Institute of Technology, Patna; JINR-Egypt: National Research Centre, Cairo; JINR-Tajikistan: TTU, PHTI, NAS RT, Dushanbe. Research on the molecular dynamics of nanoscale systems is carried out in concern with neutron scattering experiments. MD investigations are aimed at determining the structure and oscillating spectra of molecular complexes, revealing the molecular mechanisms of protein interaction, dimerization and functional characteristics of supramolecular structures and molecular complexes, establishment of the conformity to the principle and relationships of structural characteristics and functions of proteins, protein complexes and membrane-protein aggregates, analysis of the impact of composition and external parameters on the phase state of membranes, determination of structural characteristics and diffusion properties of lipid nanosystems for the transport of drugs (nanodrugs).
One of our recent achievements in the application of computer MD simulation technology concerns the efficient approach of Coarse-Grained MD calculations (CG-MD) used to simulate the structural behavior of large vesicles (biological membranes). In this area, we have been awarded the 1st Prize of the Frank Laboratory of Neutron Physics in the section "Research in Condensed Matter Physics" for the series of works "Structural reorganization of the lipid membrane induced by beta-amyloid peptide Aβ". (Authors from FLNP: A. I. Ivankov, N. Kucherka, T. N. Murugova, D. V. Soloviev, E. V. Ermakova, T. Kondela, A. V. Rogachev, A. I. Kuklin, P. Grubovchak, V. V. Skoy, Kh. T. Kholmurodov). Computer CG-MD simulation technology can significantly increase the size and time available for simulation of biological systems, such as lipid bilayers, large vesicles, filaments, others, containing from several hundred thousand to several million particles. The structural properties of model lipid membranes (large spherical vesicles) are studied and the MD simulation data are compared with experimental ones (neutron scattering and diffraction methods; SANS, small-angle neutron scattering, small-angle neutron diffraction). It is worth noting that molecular systems have been designed and studied to relate experimental and model investigations to the nature of Alzheimer's disease mechanisms. Under certain conditions, the amyloid beta oligomer can undergo an incorrect conformational rearrangement, resulting in the transition of "normal" soluble peptides to a toxic conformation, causing the production of filamentous aggregates - large insoluble rigid fibrils that is a sign of the disease. However, already isolated misfolded peptides produced in the cell membrane possess a toxic effect that has a destructive effect on the nerve cells of the brain. With the development of modern experimental techniques, the improvement of neutron and X-ray sources, the development of extra efficient detection tools and the increase in the power of computing clusters, a new round of research on Alzheimer's disease that has an important place in biomedical research, has started. Perhaps, using neutron scattering in situ with computer simulations to take a closer look at the interaction between the cell membrane and the beta-amyloid peptide will be part of the final solution to defeating Alzheimer's disease.
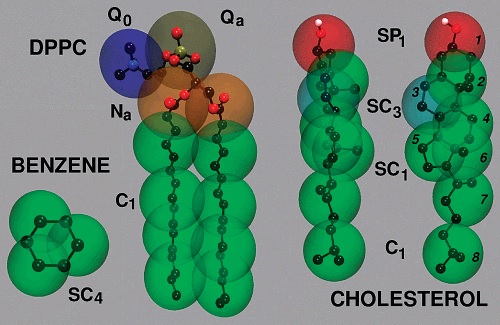
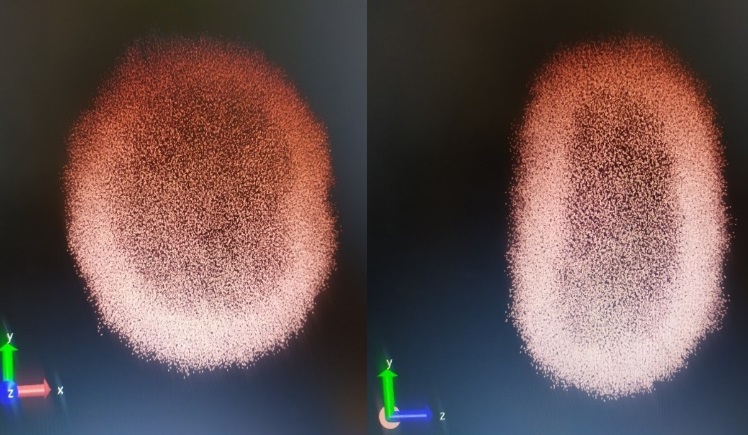
We have used the Coarse-Grained approach to Molecular Dynamics simulation of lipid vesicles in 18 Medium Amber (CG-MD). Molecular dynamics (MD) simulation of lipid vesicles of about 150 angstroms has been carried out (see below, Figures 1(a-d) and 2(a-b)). Generally, lipid-rich liposome simulation requires appropriate computational resources. Coarse-grained models of proteins, DNA, lipids can help to solve the problem of simulating large molecular systems. In this research, we have applied the CG-MD approach for lipids that uses the MORC16, MARTINI and SIRAH (http://jialuyu.com/sirah/) force fields to simulate lipid vesicles. SIRAH currently includes parameters for DNA, phospholipids and proteins (including the most common post-translational modifications) and more recently, metal ions have been included for use as cofactors. In addition, MD simulation implemented with SIRAH can take full advantage of the graphics processor acceleration and analysis software included in common MD packages.
Fig. 1 (a-d) Maps of lipid and amino acid molecules for MARTINI (a, b) and SIRAH (c, d) force field simulations using the Coarse-Grained Molecular Dynamics (CGMD).
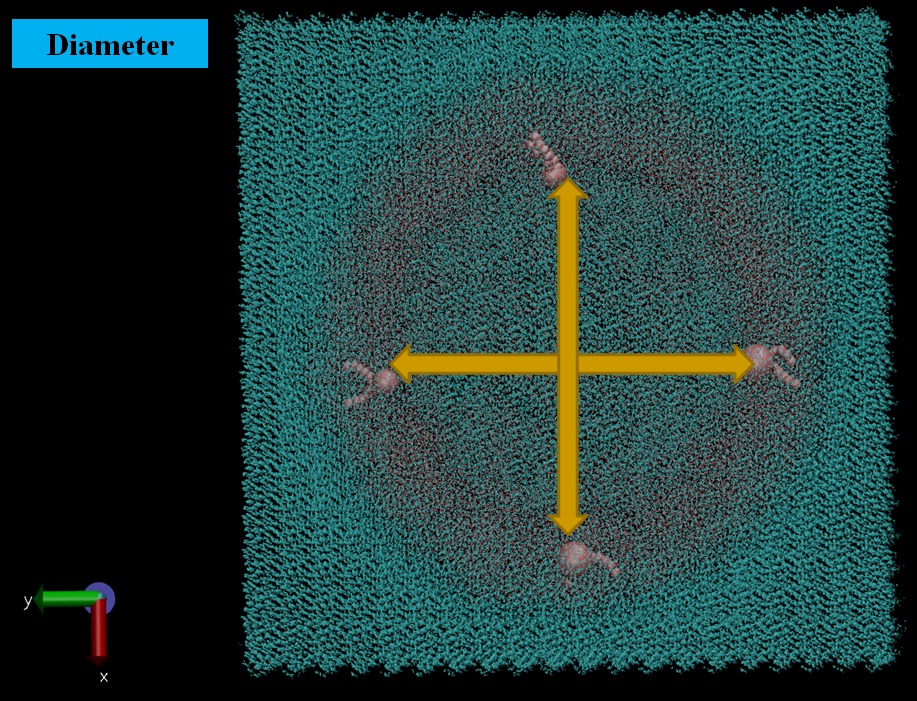
Fig. 2 (a, b) Diameter of the lipid vesicle model was evaluated as a factor of its structure stability. The initial outer diameter of the vesicle was about 150 Å. During the simulation, the maximum value of the outer diameter stabilized at about 175 Å and the minimum - at about 110 Å. These values are other factors of lipid vesicle stability.
Publications
- Pavol Hrubovčák, Ermuhammad Dushanov, Tomáš Kondela, Oleksandr Tomchuk, Kholmirzo Kholmurodov and Norbert Kučerka. (2021). Reflectometry and molecular dynamics study of the impact of cholesterol and melatonin on model lipid membranes. European Biophysics Journal, 50 (1025-1035).
- Tomáš Kondela, Ermuhammad Dushanov, Maria Vorobyeva, Kahramon Mamatkulov, Elizabeth Drolle, Dmytro Soloviov, Pavol Hrubovčák, Kholmirzo Kholmurodov, Grigory Arzumanyan, Zoya Leonenko, and Norbert Kučerka. (2021). Investigating the competitive effects of cholesterol and melatonin in model lipid membranes. BBA-Biomembranes 1863/9, 183651 (1-11).
- Norbert Kučerka, Elena Ermakova, Ermuhammad Dushanov, Kholmirzo T. Kholmurodov, Sergei Kurakin, Katarína Želinská and Daniela Uhríková. (2021) Cation-zwitterionic lipid interactions are affected by the lateral area per lipid. Langmuir 37 (278-288).
- Kučerka N., Hrubovčák P., Kondela T., Dushanov E., Kholmurodov Kh.T., Gallová J., Balgavý P. (2019) Location of the general anesthetic n-decane in model membranes. Journal of Molecular Liquids 276 (624-629).